kwarray.util_slider module¶
Defines the SlidingWindow and Sticher classes.
The SlidingWindow generates a grid of slices over an
numpy.ndarray(), which can then be used to compute on subsets of the
data. The Stitcher can then take these results and recombine them into
a final result that matches the larger array.
- class kwarray.util_slider.SlidingWindow(shape, window, overlap=None, stride=None, keepbound=False, allow_overshoot=False)[source]¶
Bases:
NiceReprSlide a window of a certain shape over an array with a larger shape.
This can be used for iterating over a grid of sub-regions of 2d-images, 3d-volumes, or any n-dimensional array.
Yields slices of shape window that can be used to index into an array with shape shape via numpy / torch fancy indexing. This allows for fast fast iteration over subregions of a larger image. Because we generate a grid-basis using only shapes, the larger image does not need to be in memory as long as its width/height/depth/etc…
- Parameters:
shape (Tuple[int, …]) – shape of source array to slide across.
window (Tuple[int, …]) – shape of window that will be slid over the larger image.
overlap (float, default=0) – a number between 0 and 1 indicating the fraction of overlap that parts will have. Specifying this is mutually exclusive with stride. Must be 0 <= overlap < 1.
stride (int, default=None) – the number of cells (pixels) moved on each step of the window. Mutually exclusive with overlap.
keepbound (bool, default=False) – if True, a non-uniform stride will be taken to ensure that the right / bottom of the image is returned as a slice if needed. Such a slice will not obey the overlap constraints. (Defaults to False)
allow_overshoot (bool, default=False) – if False, we will raise an error if the window doesn’t slide perfectly over the input shape.
- Variables:
strides (basis_shape - shape of the grid corresponding to the number of) – the sliding window will take.
dimension (basis_slices - slices that will be taken in every) –
- Yields:
Tuple[slice, …] –
- slices used for numpy indexing, the number of slices
in the tuple
Note
For each dimension, we generate a basis (which defines a grid), and we slide over that basis.
Todo
- [ ] have an option that is allowed to go outside of the window bounds
on the right and bottom when the slider overshoots.
Example
>>> from kwarray.util_slider import * # NOQA >>> shape = (10, 10) >>> window = (5, 5) >>> self = SlidingWindow(shape, window) >>> for i, index in enumerate(self): >>> print('i={}, index={}'.format(i, index)) i=0, index=(slice(0, 5, None), slice(0, 5, None)) i=1, index=(slice(0, 5, None), slice(5, 10, None)) i=2, index=(slice(5, 10, None), slice(0, 5, None)) i=3, index=(slice(5, 10, None), slice(5, 10, None))
Example
>>> from kwarray.util_slider import * # NOQA >>> shape = (16, 16) >>> window = (4, 4) >>> self = SlidingWindow(shape, window, overlap=(.5, .25)) >>> print('self.stride = {!r}'.format(self.stride)) self.stride = [2, 3] >>> list(ub.chunks(self.grid, 5)) [[(0, 0), (0, 1), (0, 2), (0, 3), (0, 4)], [(1, 0), (1, 1), (1, 2), (1, 3), (1, 4)], [(2, 0), (2, 1), (2, 2), (2, 3), (2, 4)], [(3, 0), (3, 1), (3, 2), (3, 3), (3, 4)], [(4, 0), (4, 1), (4, 2), (4, 3), (4, 4)], [(5, 0), (5, 1), (5, 2), (5, 3), (5, 4)], [(6, 0), (6, 1), (6, 2), (6, 3), (6, 4)]]
Example
>>> # Test shapes that dont fit >>> # When the window is bigger than the shape, the left-aligned slices >>> # are returend. >>> self = SlidingWindow((3, 3), (12, 12), allow_overshoot=True, keepbound=True) >>> print(list(self)) [(slice(0, 12, None), slice(0, 12, None))] >>> print(list(SlidingWindow((3, 3), None, allow_overshoot=True, keepbound=True))) [(slice(0, 3, None), slice(0, 3, None))] >>> print(list(SlidingWindow((3, 3), (None, 2), allow_overshoot=True, keepbound=True))) [(slice(0, 3, None), slice(0, 2, None)), (slice(0, 3, None), slice(1, 3, None))]
- _compute_stride(overlap, stride, shape, window)[source]¶
Ensures that stride hasoverlap the correct shape. If stride is not provided, compute stride from desired overlap.
- property grid¶
Generate indices into the “basis” slice for each dimension. This enumerates the nd indices of the grid.
- Yields:
Tuple[int, …]
- property slices¶
Generate slices for each window (equivalent to iter(self))
Example
>>> shape = (220, 220) >>> window = (10, 10) >>> self = SlidingWindow(shape, window, stride=5) >>> list(self)[41:45] [(slice(0, 10, None), slice(205, 215, None)), (slice(0, 10, None), slice(210, 220, None)), (slice(5, 15, None), slice(0, 10, None)), (slice(5, 15, None), slice(5, 15, None))] >>> print('self.overlap = {!r}'.format(self.overlap)) self.overlap = [0.5, 0.5]
- property centers¶
Generate centers of each window
- Yields:
Tuple[float, …] – the center coordinate of the slice
Example
>>> shape = (4, 4) >>> window = (3, 3) >>> self = SlidingWindow(shape, window, stride=1) >>> list(zip(self.centers, self.slices)) [((1.0, 1.0), (slice(0, 3, None), slice(0, 3, None))), ((1.0, 2.0), (slice(0, 3, None), slice(1, 4, None))), ((2.0, 1.0), (slice(1, 4, None), slice(0, 3, None))), ((2.0, 2.0), (slice(1, 4, None), slice(1, 4, None)))] >>> shape = (3, 3) >>> window = (2, 2) >>> self = SlidingWindow(shape, window, stride=1) >>> list(zip(self.centers, self.slices)) [((0.5, 0.5), (slice(0, 2, None), slice(0, 2, None))), ((0.5, 1.5), (slice(0, 2, None), slice(1, 3, None))), ((1.5, 0.5), (slice(1, 3, None), slice(0, 2, None))), ((1.5, 1.5), (slice(1, 3, None), slice(1, 3, None)))]
- class kwarray.util_slider.Stitcher(shape, device='numpy', dtype='float32', nan_policy='propogate')[source]¶
Bases:
NiceReprStitches multiple possibly overlapping slices into a larger array.
This is used to invert the SlidingWindow. For semenatic segmentation the patches are probability chips. Overlapping chips are averaged together.
- SeeAlso:
kwarray.RunningStats- similarly performs running means, butcan also track other statistics.
Example
>>> from kwarray.util_slider import * # NOQA >>> import sys >>> # Build a high resolution image and slice it into chips >>> highres = np.random.rand(5, 200, 200).astype(np.float32) >>> target_shape = (1, 50, 50) >>> slider = SlidingWindow(highres.shape, target_shape, overlap=(0, .5, .5)) >>> # Show how Sticher can be used to reconstruct the original image >>> stitcher = Stitcher(slider.input_shape) >>> for sl in list(slider): ... chip = highres[sl] ... stitcher.add(sl, chip) >>> assert stitcher.weights.max() == 4, 'some parts should be processed 4 times' >>> recon = stitcher.finalize()
Example
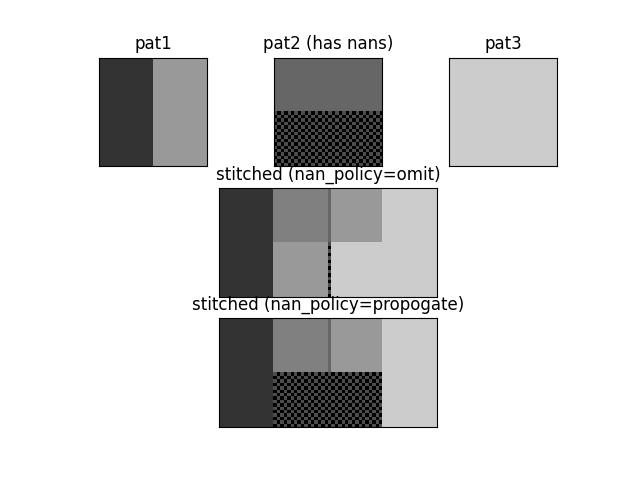
>>> from kwarray.util_slider import * # NOQA >>> import sys >>> # Demo stitching 3 patterns where one has nans >>> pat1 = np.full((32, 32), fill_value=0.2) >>> pat2 = np.full((32, 32), fill_value=0.4) >>> pat3 = np.full((32, 32), fill_value=0.8) >>> pat1[:, 16:] = 0.6 >>> pat2[16:, :] = np.nan >>> # Test with nan_policy=omit >>> stitcher = Stitcher(shape=(32, 64), nan_policy='omit') >>> stitcher[0:32, 0:32](pat1) >>> stitcher[0:32, 16:48](pat2) >>> stitcher[0:32, 33:64](pat3[:, 1:]) >>> final1 = stitcher.finalize() >>> # Test without nan_policy=propogate >>> stitcher = Stitcher(shape=(32, 64), nan_policy='propogate') >>> stitcher[0:32, 0:32](pat1) >>> stitcher[0:32, 16:48](pat2) >>> stitcher[0:32, 33:64](pat3[:, 1:]) >>> final2 = stitcher.finalize() >>> # Checks >>> assert np.isnan(final1).sum() == 16, 'only should contain nan where no data was stiched' >>> assert np.isnan(final2).sum() == 512, 'should contain nan wherever a nan was stitched' >>> # xdoctest: +REQUIRES(--show) >>> # xdoctest: +REQUIRES(module:kwplot) >>> import kwplot >>> import kwimage >>> kwplot.autompl() >>> kwplot.imshow(pat1, title='pat1', pnum=(3, 3, 1)) >>> kwplot.imshow(kwimage.nodata_checkerboard(pat2, square_shape=1), title='pat2 (has nans)', pnum=(3, 3, 2)) >>> kwplot.imshow(pat3, title='pat3', pnum=(3, 3, 3)) >>> kwplot.imshow(kwimage.nodata_checkerboard(final1, square_shape=1), title='stitched (nan_policy=omit)', pnum=(3, 1, 2)) >>> kwplot.imshow(kwimage.nodata_checkerboard(final2, square_shape=1), title='stitched (nan_policy=propogate)', pnum=(3, 1, 3))
Example
>>> # Example of weighted stitching >>> # xdoctest: +REQUIRES(module:kwimage) >>> from kwarray.util_slider import * # NOQA >>> import kwimage >>> import kwarray >>> import sys >>> data = kwimage.Mask.demo().data.astype(np.float32) >>> data_dims = data.shape >>> window_dims = (8, 8) >>> # We are going to slide a window over the data, do some processing >>> # and then stitch it all back together. There are a few ways we >>> # can do it. Lets demo the params. >>> basis = { >>> # Vary the overlap of the slider >>> 'overlap': (0, 0.5, .9), >>> # Vary if we are using weighted stitching or not >>> 'weighted': ['none', 'gauss'], >>> 'keepbound': [True, False] >>> } >>> results = [] >>> gauss_weights = kwimage.gaussian_patch(window_dims) >>> gauss_weights = kwimage.normalize(gauss_weights) >>> for params in ub.named_product(basis): >>> if params['weighted'] == 'none': >>> weights = None >>> elif params['weighted'] == 'gauss': >>> weights = gauss_weights >>> # Build the slider and stitcher >>> slider = kwarray.SlidingWindow( >>> data_dims, window_dims, overlap=params['overlap'], >>> allow_overshoot=True, >>> keepbound=params['keepbound']) >>> stitcher = kwarray.Stitcher(data_dims) >>> # Loop over the regions >>> for sl in list(slider): >>> chip = data[sl] >>> # This is our dummy function for thie example. >>> predicted = np.ones_like(chip) * chip.sum() / chip.size >>> stitcher.add(sl, predicted, weight=weights) >>> final = stitcher.finalize() >>> results.append({ >>> 'final': final, >>> 'params': params, >>> }) >>> # xdoctest: +REQUIRES(--show) >>> # xdoctest: +REQUIRES(module:kwplot) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nCols=3, nSubplots=len(results) + 2) >>> kwplot.imshow(data, pnum=pnum_(), title='input image') >>> kwplot.imshow(gauss_weights, pnum=pnum_(), title='Gaussian weights') >>> pnum_() >>> for result in results: >>> param_key = ub.urepr(result['params'], compact=1) >>> final = result['final'] >>> canvas = kwarray.normalize(final) >>> canvas = kwimage.fill_nans_with_checkers(canvas) >>> kwplot.imshow(canvas, pnum=pnum_(), title=param_key)

- Parameters:
shape (tuple) – dimensions of the large image that will be created from the smaller pixels or patches.
device (str | int | torch.device) – default is ‘numpy’, but if given as a torch device, then underlying operations will be done with torch tensors instead.
dtype (str) – the datatype to use in the underlying accumulator.
nan_policy (str) – if omit, check for nans and convert any to zero weight items in stitching.
- add(indices, patch, weight=None)[source]¶
Incorporate a new (possibly overlapping) patch or pixel using a weighted sum.
- Parameters:
indices (slice | tuple | None) – typically a Tuple[slice] of pixels or a single pixel, but this can be any numpy fancy index.
patch (ndarray) – data to patch into the bigger image.
weight (float | ndarray) – weight of this patch (default to 1.0)
- kwarray.util_slider._slices1d(margin, stop, step=None, start=0, keepbound=False, check=True)[source]¶
Helper to generates slices in a single dimension.
- Parameters:
margin (int) – the length of the slice (window)
stop (int) – the length of the image dimension
step (int, default=None) – the length of each step / distance between slices
start (int, default=0) – starting point (in most cases set this to 0)
keepbound (bool) – if True, a non-uniform step will be taken to ensure that the right / bottom of the image is returned as a slice if needed. Such a slice will not obey the overlap constraints. (Defaults to False)
check (bool) – if True an error will be raised if the window does not cover the entire extent from start to stop, even if keepbound is True.
- Yields:
slice – slice in one dimension of size (margin)
Example
>>> stop, margin, step = 2000, 360, 360 >>> keepbound = True >>> strides = list(_slices1d(margin, stop, step, keepbound, check=False)) >>> assert all([(s.stop - s.start) == margin for s in strides])
Example
>>> stop, margin, step = 200, 46, 7 >>> keepbound = True >>> strides = list(_slices1d(margin, stop, step, keepbound=False, check=True)) >>> starts = np.array([s.start for s in strides]) >>> stops = np.array([s.stop for s in strides]) >>> widths = stops - starts >>> assert np.all(np.diff(starts) == step) >>> assert np.all(widths == margin)
Example
>>> import pytest >>> stop, margin, step = 200, 36, 7 >>> with pytest.raises(ValueError): ... list(_slices1d(margin, stop, step))